The integrated microbial genomes (IMG) system serves as a neighborhood useful resource for comparative analysis of publicly obtainable genomes in a complete integrated context.
IMG accommodates each draft and full microbial genomes integrated with different publicly obtainable genomes from all three domains of life, along with a lot of plasmids and viruses. IMG supplies instruments and viewers for analyzing and reviewing the annotations of genes and genomes in a comparative context.
Since its first launch in 2005, IMG’s knowledge content material and analytical capabilities have been continually expanded via common releases. Several companion IMG techniques have been arrange with a view to serve area particular wants, reminiscent of skilled evaluation of genome annotation
Ten years of sustaining and expanding a microbial genome and metagenome analysis system.
Launched in March 2005, the Integrated Microbial Genomes (IMG) system is a complete knowledge administration system that helps multidimensional comparative analysis of genomic knowledge.
At the core of the IMG system is a knowledge warehouse that accommodates genome and metagenome datasets sequenced on the Joint Genome Institute or supplied by scientific customers, in addition to public genome datasets obtainable on the National Center for Biotechnology Information Genbank sequence knowledge archive.
Genomes and metagenome datasets are processed utilizing IMG’s microbial genome and metagenome sequence knowledge processing pipelines and are integrated into the info warehouse utilizing IMG’s knowledge integration toolkits. Microbial genome and metagenome utility particular knowledge marts and person interfaces present entry to completely different subsets of IMG’s knowledge and analysis toolkits.
This evaluation article revisits IMG’s authentic goals, highlights key milestones reached by the system through the previous 10 years, and discusses the primary challenges confronted by a quickly expanding system, particularly the complexity of sustaining such a system in an educational setting with restricted budgets and computing and knowledge administration infrastructure.
Comparative genome analysis within the integrated microbial genomes (IMG) system.
Comparative genome analysis is crucial for the efficient exploration of a quickly rising variety of full and draft sequences for microbial genomes.
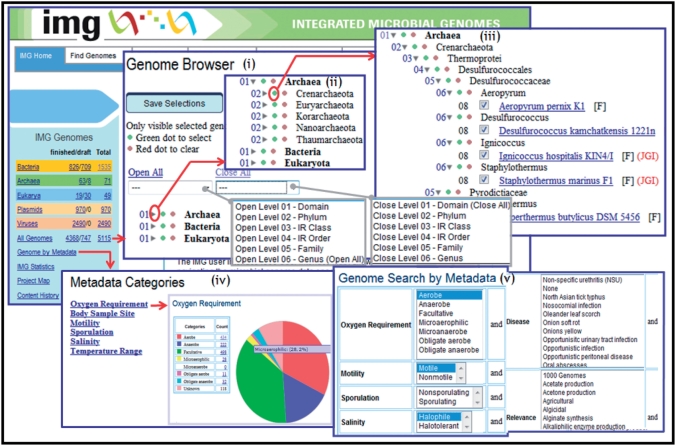
The Integrated Microbial Genomes (IMG) system (img.jgi.doe.gov) has been developed as a neighborhood useful resource that gives assist for comparative analysis of microbial genomes in an integrated context. IMG permits customers to navigate the multidimensional microbial genome knowledge area and focus their analysis on a subset of genes, genomes, and capabilities of curiosity.
IMG supplies graphical viewers, summaries, and prevalence profile instruments for evaluating genes, pathways, and capabilities (phrases) throughout particular genomes. Genes might be additional examined utilizing gene neighborhoods and in contrast with sequence alignment instruments.
